-
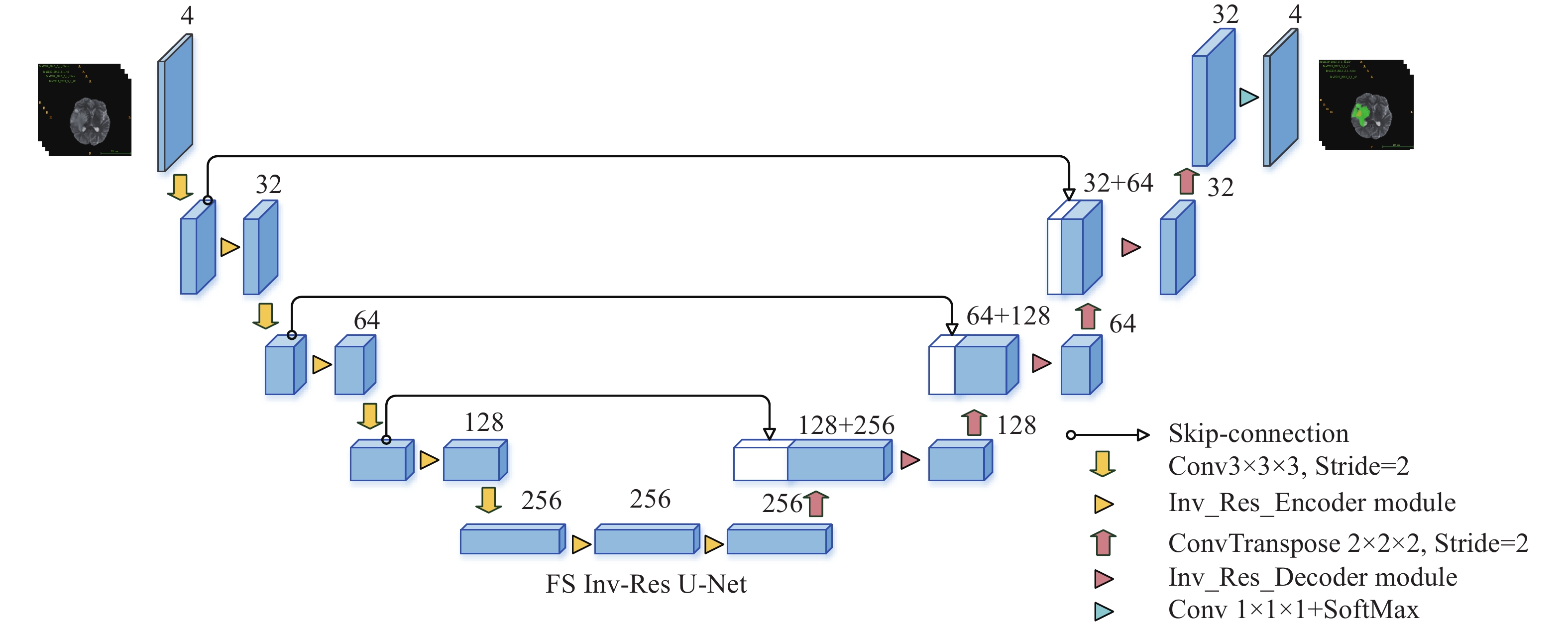
摘要: 近年來卷積神經網絡在生物醫學圖像處理中得到了廣泛應用,例如從磁共振圖像中準確分割腦腫瘤是臨床診斷和治療腦部腫瘤疾病的關鍵環節。3D U-Net因其分割效果優異受到追捧,但其跳躍連接補充的特征圖為編碼器特征提取后的輸出特征圖,并未進一步考慮到此過程中的原始細節信息丟失問題。針對這一問題,本文提出前置跳躍連接,并在此基礎上設計了一種前置跳躍連接倒殘差U形網絡(FS Inv-Res U-Net)。首先,將前置跳躍連接用于改進DMF Net、HDC Net和3D U-Net 3個典型網絡以驗證其有效性和泛化性;其次,采用前置跳躍連接和倒殘差結構改進3D U-Net,進而提出FS Inv-Res U-Net,最后在BraTS公開驗證集上對所提網絡進行驗證。BraTS2018的驗證結果在增強型腫瘤、全腫瘤和腫瘤核心的Dice值分別是80.23%、90.30%和85.45%,豪斯多夫距離分別是2.35、4.77和5.50 mm;BraTS2019的驗證結果在增強型腫瘤、全腫瘤和腫瘤核心的Dice值分別是78.38%、89.78%和83.01%,豪斯多夫距離分別是4、5.57和6.37 mm。結果表明,FS Inv-Res U-Net取得了不輸于先進網絡的評價指標,能夠實現腦腫瘤精確分割。Abstract: Accurate segmentation of brain tumors from magnetic resonance images is the key to the clinical diagnosis and rational treatment of brain tumor diseases. Recently, convolutional neural networks have been widely used in biomedical image processing. 3D U-Net is sought after because of its excellent segmentation effect; however, the feature map supplemented by the skip connection is the output feature map after the encoder feature extraction, and the loss of original detail information in this process is ignored. In the 3D U-Net design, after each layer of convolution, regularization, and activation function processing, the detailed information contained in the feature map will deviate from the original detailed information. For skip connections, the essence of this design is to supplement the detailed information of the original features to the decoder; that is, in the decoder stage, the more original the skip connection-supplemented feature maps are, the more easily the decoder can achieve a better segmentation effect. To address this problem, this paper proposes the concept of a front-skip connection. That is, the starting point of the skip connection is adjusted to the front to improve the network performance. On the basis of this idea, we design a front-skip connection inverted residual U-shaped network (FS Inv-Res U-Net). First, the front-skip connections are applied to three typical networks, DMF Net, HDC Net, and 3D U-Net, to verify their effectiveness and generalization. Applying our proposed front-skip connection concept on these three networks improves the network performance, indicating that the idea of a front-skip connection is simple but powerful and has out-of-the-box characteristics. Second, 3D U-Net is enhanced using the front-skip connection concept and the inverted residual structure of MobileNet, and then FS Inv-Res U-Net is proposed based on these two ideas. Additionally, ablation experiments are conducted on FS Inv-Res U-Net. After adding the front-skip connection and the inverted residual module to the backbone network 3D U-Net, the segmentation performance of the proposed network is greatly improved, indicating that the front-skip connection and the inverted residual module help our brain tumor segmentation network. Finally, the proposed network is validated on the validation dataset of the public datasets BraTS 2018 and BraTS 2019. The Dice scores of the validation results on the enhanced tumor, whole tumor, and tumor core were 80.23%, 90.30%, and 85.45% and 78.38%, 89.78%, and 83.01%, respectively; the hausdorff95 distances were 2.35, 4.77, and 5.50 mm and 4, 5.57, and 6.37 mm, respectively. The above results show that the FS Inv-Res U-Net proposed in this paper achieves the same evaluation indicators as advanced networks and provides accurate brain tumor segmentations.
-
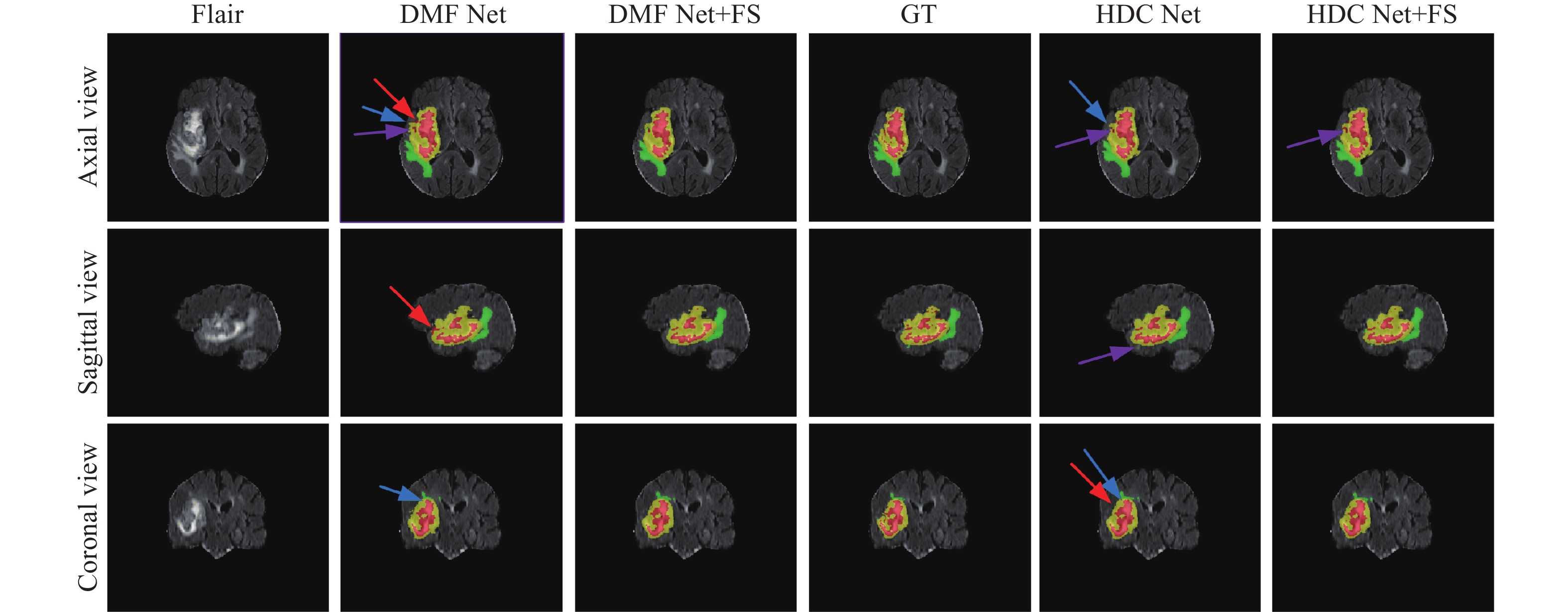
圖 3 前置跳躍連接的消融實驗(FS 表示前置跳躍連接,GT表示標準真值,紅色箭頭表示錯誤分類區域,藍色箭頭表示邊界信息丟失,紫色箭頭表示子區域與真值區域大小不同)
Figure 3. Ablation experiment of front-skip connections (FS indicates pre-skip connections, GT indicates ground truth, red arrows indicate misclassified regions, blue arrows indicate loss of boundary information, and purple arrows indicate subregions with different sizes than ground truth regions)
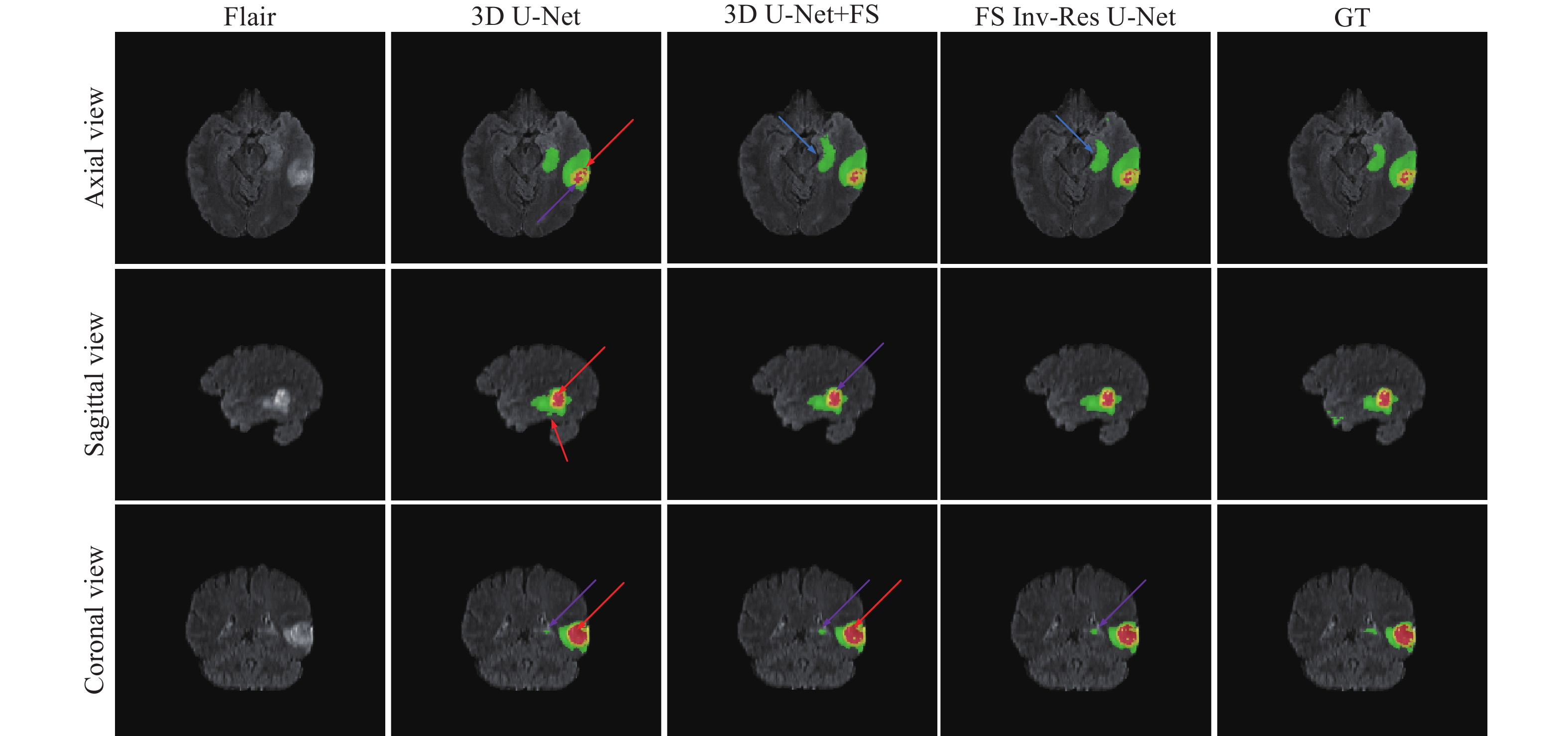
圖 4 FS Inv-Res U-Net的消融實驗(FS表示前置跳躍連接,GT表示標準真值,紅色箭頭表示錯誤分類區域,藍色箭頭表示邊界信息丟失,紫色箭頭表示子區域與真值區域大小不同)
Figure 4. Ablation experiments of FS Inv-Res U-Net (FS indicates the front-skip connection, GT indicates the ground truth, red arrows indicate misclassified regions, blue arrows indicate loss of boundary information, and purple arrows indicate subregions with different sizes than ground truth regions)
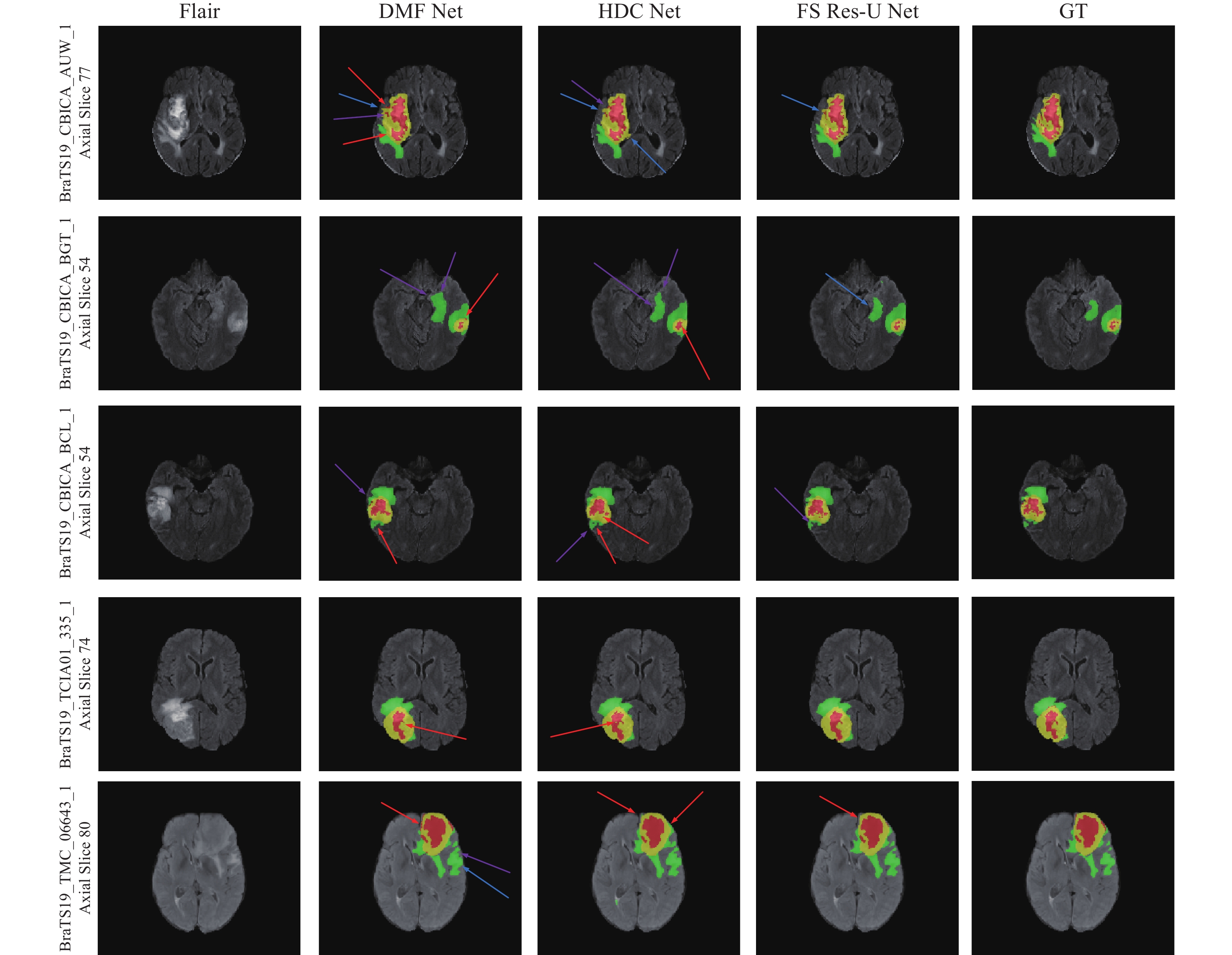
圖 5 不同方法的分割圖對比(紅色箭頭表示錯誤分類區域,藍色箭頭表示邊界信息丟失,紫色箭頭表示子區域與真值區域大小不同)
Figure 5. Comparison of different methods’ segmentation maps (Red arrows indicate misclassified regions, blue arrows indicate loss of boundary information, and purple arrows indicate subregions with different sizes than ground truth regions)
表 1 前置跳躍連接的消融實驗
Table 1. Ablation experiments of front-skip connections
BraTS Methods Dice/% Hausdorff95/mm ET WT TC AVG ET WT TC AVG 2018 DMF Net[23] 80.12 90.62 84.54 85.09 3.06 4.66 6.44 4.72 DMF Net+FS 80.85 90.36 84.64 85.28 2.73 3.92 6.71 4.45 HDC Net[24] 80.90 89.70 84.70 85.10 2.43 4.62 6.12 4.39 HDC Net+FS 80.94 89.98 85.04 85.32 2.33 4.19 5.60 4.04 3D U-Net[13] 80.06 89.86 83.98 84.63 2.41 4.62 5.95 4.33 3D U-Net+FS 80.14 90.15 85.15 85.15 2.56 5.88 5.57 4.67 2019 DMF Net[23] 77.33 89.75 81.67 82.92 2.84 6.19 6.59 5.21 DMF Net+FS 77.54 89.82 81.93 83.10 2.53 4.65 6.33 4.50 HDC Net[24] 77.35 89.30 81.83 82.83 4.19 6.74 7.98 6.30 HDC Net+FS 77.37 89.78 82.89 83.35 3.01 5.66 5.58 4.75 3D U-Net[13] 77.83 89.39 81.72 82.98 4.23 6.71 6.87 5.94 3D U-Net+FS 78.32 89.82 82.50 83.55 4.27 5.40 6.68 5.45 Notes: FS indicates the front-skip connection, AVG indicates the average value of three subregions, and the bold numbers in the table indicate the optimal value of the same area index for the same network. 表 2 FS Inv-Res U-Net的消融實驗
Table 2. Ablation experiments of FS Inv-Res U-Net
BraTS Methods Dice/% Hausdorff95/mm ET WT TC AVG ET WT TC AVG 2018 3D U-Net[13] 80.06 89.86 83.98 84.63 2.41 4.62 5.95 4.33 3D U-Net+FS 80.14 90.15 85.15 85.15 2.56 5.88 5.57 4.67 FS Inv-Res U-Net 80.23 90.30 85.45 85.33 2.35 4.77 5.50 4.21 2019 3D U-Net[13] 77.83 89.39 81.72 82.98 4.23 6.71 6.87 5.94 3D U-Net+FS 78.32 89.82 82.50 83.55 4.27 5.40 6.68 5.45 FS Inv-Res U-Net 78.38 89.78 83.01 83.72 4.00 5.57 6.37 5.31 Notes: FS indicates front-skip connection, the bold numbers in the table represent the optimal value of the index in the same region, and the underlined numbers indicate the second optimal value of the index in the same area. 表 3 FS Inv-Res U-Net與其他先進網絡的對比
Table 3. Comparison of FS Inv-Res U-Net with other state-of-the-art networks
BraTS Methods Dice/% Hausdorff95/mm ET WT TC AVG ET WT TC AVG 2018 MSMA Net[28] 75.80 89.00 81.10 81.97 5.06 13.6 10.6 9.75 CA Net[29] 76.70 89.80 83.40 83.30 3.86 6.69 7.67 6.07 AE AU-Net[30] 80.00 90.8 83.8 84.87 2.59 4.55 8.14 5.09 DMF Net[23] 80.12 90.62 84.54 85.09 3.06 4.66 6.44 4.72 HDC Net[24] 80.90 89.70 84.70 85.10 2.43 4.62 6.12 4.39 NVDLMED[31] 81.73 90.68 86.02 86.14 3.82 4.52 6.85 5.06 This work 80.23 90.30 85.45 85.33 2.35 4.77 5.50 4.21 2019 Zhou et al.[32] 72.70 87.10 71.80 77.20 6.30 6.70 9.30 7.40 KiU-Net 3D[33] 73.21 87.60 73.92 78.24 6.32 8.94 9.89 8.38 SoResU-Net[34] 72.40 87.50 87.50 82.47 5.97 9.35 11.47 8.93 HDC Net[24] 77.35 89.30 81.83 82.83 4.19 6.74 7.98 6.30 DMF Net[23] 77.33 89.75 81.67 82.92 2.84 6.19 6.59 5.21 AE AU-Net[30] 77.30 90.20 81.50 83.00 4.65 6.15 7.54 6.11 CA Net[29] 75.90 88.50 85.10 83.17 4.81 7.09 8.41 6.77 Zhao et al.[35] 75.40 91.00 83.50 83.30 3.84 4.57 5.58 4.66 This work 78.38 89.78 83.01 83.72 4.00 5.57 6.37 5.31 Notes: The numbers in bold indicate the optimal values of the same area. www.77susu.com<span id="fpn9h"><noframes id="fpn9h"><span id="fpn9h"></span> <span id="fpn9h"><noframes id="fpn9h"> <th id="fpn9h"></th> <strike id="fpn9h"><noframes id="fpn9h"><strike id="fpn9h"></strike> <th id="fpn9h"><noframes id="fpn9h"> <span id="fpn9h"><video id="fpn9h"></video></span> <ruby id="fpn9h"></ruby> <strike id="fpn9h"><noframes id="fpn9h"><span id="fpn9h"></span> -
參考文獻
[1] Cong M, Wu T, Liu D, et al. Prostate MR/TRUS image segmentation and registration methods based on supervised learning. Chin J Eng, 2020, 42(10): 1362叢明, 吳童, 劉冬, 等. 基于監督學習的前列腺MR/TRUS圖像分割和配準方法. 工程科學學報, 2020, 42(10):1362 [2] Chen B S, Zhang L L, Chen H Y, et al. A novel extended kalman filter with support vector machine based method for the automatic diagnosis and segmentation of brain tumors. Comput Meth Prog Bio, 2021, 200: 105797 doi: 10.1016/j.cmpb.2020.105797 [3] Pinto A, Pereira S, Rasteiro D, et al. Hierarchical brain tumour segmentation using extremely randomized trees. Pattern Recogn, 2018, 82: 105 doi: 10.1016/j.patcog.2018.05.006 [4] Chen G X, Li Q, Shi F Q, et al. RFDCR: Automated brain lesion segmentation using cascaded random forests with dense conditional random fields. NeuroImage, 2020, 211: 116620 doi: 10.1016/j.neuroimage.2020.116620 [5] Jiang D G, Li M M, Chen Y Z, et al. Cascaded retinal vessel segmentation network guided by a skeleton map. Chin J Eng, 2021, 43(9): 1244姜大光, 李明鳴, 陳羽中, 等. 骨架圖引導的級聯視網膜血管分割網絡. 工程科學學報, 2021, 43(9):1244 [6] Liu H X, Li Q, Wang I C. A deep-learning model with learnable group convolution and deep supervision for brain tumor segmentation. Math Probl Eng, 2021, 2021: 6661083 [7] Ali M J, Raza B, Shahid A R. Multi-level kronecker convolutional neural network (ML-KCNN) for glioma segmentation from multi-modal MRI volumetric data. J Digit Imaging, 2021, 34(4): 905 doi: 10.1007/s10278-021-00486-7 [8] Cui S G, Wei M J, Liu C, et al. GAN-segNet: A deep generative adversarial segmentation network for brain tumor semantic segmentation. Int J Imaging Syst Tech, 2022, 32(3): 857 doi: 10.1002/ima.22677 [9] Cheng G H, Ji H L, He L Y. Correcting and reweighting false label masks in brain tumor segmentation. Med Phys, 2021, 48(1): 169 doi: 10.1002/mp.14480 [10] Rahimpour M, Bertels J, Vandermeulen D, et al. Improving T1w MRI-based brain tumor segmentation using cross-modal distillation // Society of Photo-Optical Instrumentation Engineers (SPIE) Conference Series. 2021, 11596: 261 [11] Jiang Z Y, Ding C X, Liu M F, et al. Two-stage cascaded U-Net: 1st place solution to BraTS challenge 2019 segmentation task // International MICCAI Brainlesion Workshop. Lima, 2020: 231 [12] Zhang L, Zhang J M, Shen P Y, et al. Block level skip connections across cascaded V-Net for multi-organ segmentation. IEEE Trans Med Imaging, 2020, 39(9): 2782 doi: 10.1109/TMI.2020.2975347 [13] ?i?ek ?, Abdulkadir A, Lienkamp S S, et al. 3D U-Net: Learning dense volumetric segmentation from sparse annotation // International Conference on Medical Image Computing and Computer-Assisted Intervention. Athens, 2016: 424 [14] Huang G, Liu Z, Maaten L, et al. Densely connected convolutional networks // 2017 IEEE Conference on Computer Vision and Pattern Recognition. Honolulu, 2017: 2261 [15] Wu J, Zhang Y, Wang K, et al. Skip connection U-Net for white matter hyperintensities segmentation from MRI. IEEE Access, 7: 155194 [16] Ma B F, Chang C Y. Semantic segmentation of high-resolution remote sensing images using multiscale skip connection network. IEEE Sens J, 2022, 22(4): 3745 doi: 10.1109/JSEN.2021.3139629 [17] Zhou Z W, Siddiquee M M R, Tajbakhsh N, et al. UNet: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans Med Imaging, 2020, 39(6): 1856 doi: 10.1109/TMI.2019.2959609 [18] He K M, Zhang X Y, Ren S Q, et al. Deep residual learning for image recognition // 2016 IEEE Conference on Computer Vision and Pattern Recognition. Las Vegas, 2016: 770 [19] Sandler M, Howard A, Zhu M L, et al. MobileNetV2: Inverted residuals and linear bottlenecks // 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. Salt Lake City, 2018: 4510 [20] Yu C G, Liu K, Zou W. A method of small object detection based on improved deep learning. Opt Mem Neural Networks, 2020, 29(2): 69 doi: 10.3103/S1060992X2002006X [21] Zhao W L, Wang Z J, Cai W A, et al. Multiscale inverted residual convolutional neural network for intelligent diagnosis of bearings under variable load condition. Measurement, 2022, 188: 110511 doi: 10.1016/j.measurement.2021.110511 [22] Zhang T Y, Shi C P, Liao D L, et al. Deep spectral spatial inverted residual network for hyperspectral image classification. Remote Sens, 2021, 13(21): 4472 doi: 10.3390/rs13214472 [23] Chen C, Liu X P, Ding M, et al. 3D dilated multi-fiber network for real-time brain tumor segmentation in MRI // International Conference on Medical Image Computing and Computer-Assisted Intervention. Shenzhen, 2019: 184 [24] Luo Z R, Jia Z D, Yuan Z M, et al. HDC-net: Hierarchical decoupled convolution network for brain tumor segmentation. IEEE J Biomed Health Inform, 2021, 25(3): 737 doi: 10.1109/JBHI.2020.2998146 [25] Sudre C H, Li W Q, Vercauteren T, et al. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations // International Workshop on Deep Learning in Medical Image Analysis & International Workshop on Multimodal Learning for Clinical Decision Support. Québec City, 2017: 240 [26] Menze B H, Jakab A, Bauer S, et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging, 2015, 34(10): 1993 doi: 10.1109/TMI.2014.2377694 [27] Bakas S, Akbari H, Sotiras A, et al. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data, 2017, 4(1): 1 [28] Zhang Y, Lu Y, Chen W K, et al. MSMANet: A multi-scale mesh aggregation network for brain tumor segmentation. Appl Soft Comput, 2021, 110: 107733 doi: 10.1016/j.asoc.2021.107733 [29] Liu Z H, Tong L, Chen L, et al. CANet: Context aware network for brain glioma segmentation. IEEE Trans Med Imaging, 2021, 40(7): 1763 doi: 10.1109/TMI.2021.3065918 [30] Rosas-Gonzalez S, Birgui-Sekou T, Hidane M, et al. Asymmetric ensemble of asymmetric U-net models for brain tumor segmentation with uncertainty estimation. Front Neurol, 2021, 12: 609646 doi: 10.3389/fneur.2021.609646 [31] Myronenko A. 3D MRI brain tumor segmentation using autoencoder regularization // International MICCAI Brainlesion Workshop. Shenzhen, 2019: 311 [32] Zhou T X, Canu S, Vera P, et al. Latent correlation representation learning for brain tumor segmentation with missing MRI modalities. IEEE Trans Image Process, 2021, 30: 4263 doi: 10.1109/TIP.2021.3070752 [33] Valanarasu J M J, Sindagi V A, Hacihaliloglu I, et al. KiU-net: Overcomplete convolutional architectures for biomedical image and volumetric segmentation. IEEE Trans Med Imaging, 2022, 41(4): 965 doi: 10.1109/TMI.2021.3130469 [34] Sheng N, Liu D W, Zhang J X, et al. Second-order ResU-Net for automatic MRI brain tumor segmentation. Math Biosci Eng, 2021, 18(5): 4943 doi: 10.3934/mbe.2021251 [35] Zhao Y X, Zhang Y M, Liu C L. Bag of tricks for 3D MRI brain tumor segmentation // International MICCAI Brainlesion Workshop. Lima, 2020: 210 -



 下載:
下載: